Note
Go to the end to download the full example code.
Interpolation on Graphene deflector scan¶
Simple workflow for the interpolation of data along a generic path in the k-space from its isoenergy cuts. The data are a deflector scan on graphene as simulated from a third nearest neighbor tight binding model. The same workflow can be applied to any tilt-, polar-, deflector- or hv-scan.
Import the “fundamental” python libraries for a generic data analysis:
import numpy as np
import matplotlib.pyplot as plt
Instead of loading the file as for example:
# from navarp.utils import navfile
# file_name = r"nxarpes_simulated_cone.nxs"
# entry = navfile.load(file_name)
Here we build the simulated graphene signal with a dedicated function defined just for this purpose:
from navarp.extras.simulation import get_tbgraphene_deflector
entry = get_tbgraphene_deflector(
scans=np.linspace(-5., 20., 91),
angles=np.linspace(-25, 6, 400),
ebins=np.linspace(-13, 0.4, 700),
tht_an=-18,
phi_an=0,
hv=120,
gamma=0.05
)
Fermi level autoset¶
entry.autoset_efermi(scan_range=[-5, 5], energy_range=[115.2, 115.8])
print("Energy of the Fermi level = {:.0f} eV".format(entry.efermi))
print("Energy resolution = {:.0f} meV".format(entry.efermi_fwhm*1000))
entry.plt_efermi_fit()
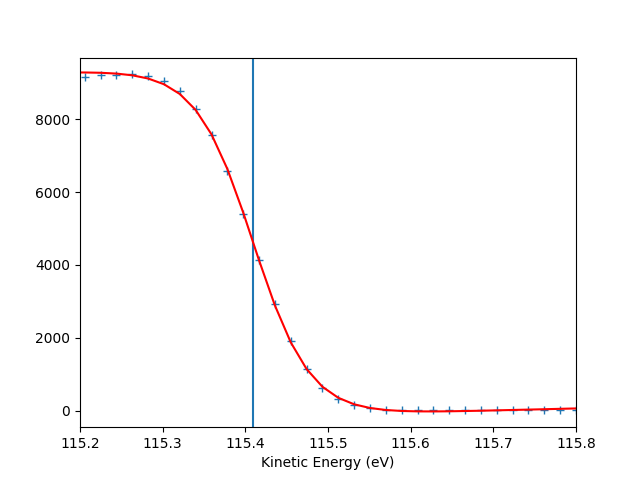
Fermi level at 115.4093 eV
Energy resolution = 137.9 meV (i.e. FWHM of the Gaussian shape which, convoluted with a step function, fits the Fermi edge)
Photon energy is now set to 120.0093 eV (instead of 120.0000 eV)
Energy of the Fermi level = 115 eV
Energy resolution = 138 meV
Check for the Fermi level alignment¶
entry.isoscan(scan=0, dscan=0).show(yname='eef')
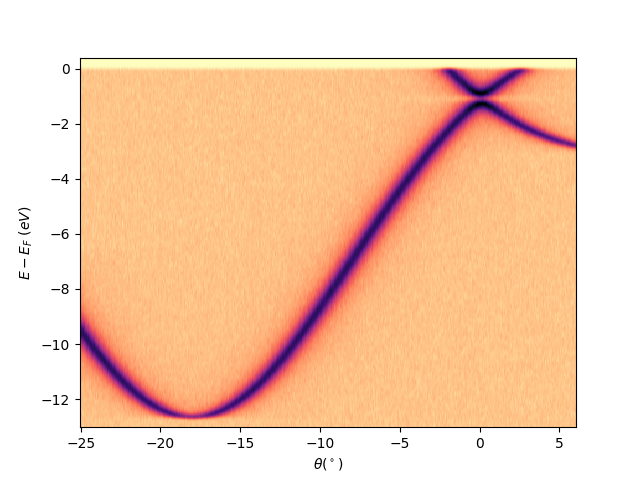
<matplotlib.collections.QuadMesh object at 0x78b8ea971e40>
Plotting iso-energetic cut at ekin = efermi¶
entry.isoenergy(0, 0.02).show()
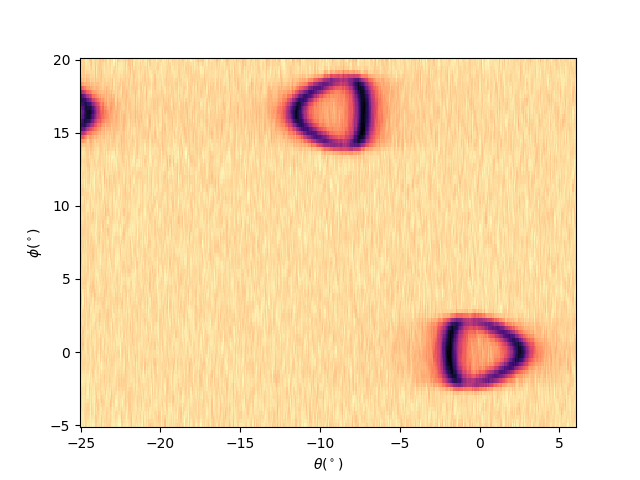
<matplotlib.collections.QuadMesh object at 0x78b8ea8f5660>
Set the k-space for the transformation¶
entry.set_kspace(
tht_p=0.1,
k_along_slit_p=1.7,
scan_p=0,
ks_p=0,
e_kin_p=114.3,
)
tht_an = -17.979
scan_type = deflector
inn_pot = 14.000
scans_0 = 0.000
phi_an = 0.000
kspace transformation ready
and check the isoenergy at the Fermi level:
entry.isoenergy(0, 0.02).show()
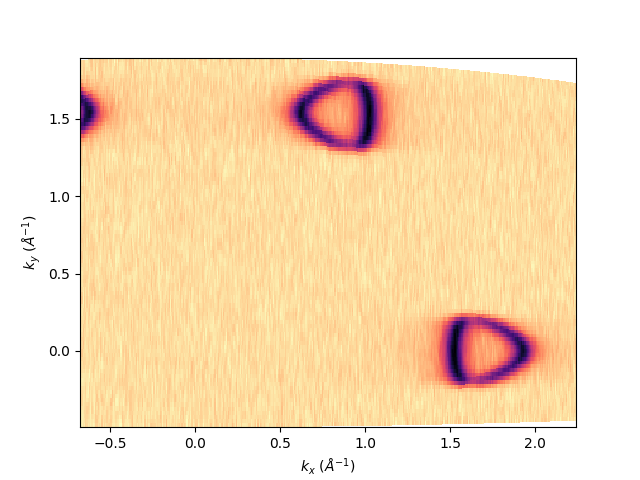
<matplotlib.collections.QuadMesh object at 0x78b8eaf4f370>
Define the interpolation path points¶
kbins = 900
k_GK = 1.702
k_pts_xy = np.array([
[0, 0],
[k_GK, 0],
[k_GK*np.cos(np.pi/3), k_GK*np.sin(np.pi/3)+0.05],
[0, 0]
])
kx_pts = k_pts_xy[:, 0]
ky_pts = k_pts_xy[:, 1]
klabels = [
r'$\Gamma$',
r'$\mathrm{K}$',
r'$\mathrm{K}^{\prime}$',
r'$\Gamma$'
]
and show them on the isoenergy at the Dirac point energy:
entry.isoenergy(-1.1, 0.02).show()
plt.plot(kx_pts, ky_pts, '-+')
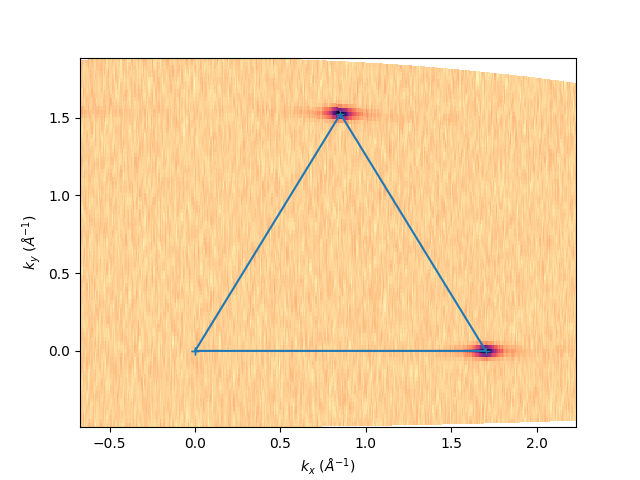
[<matplotlib.lines.Line2D object at 0x78b8eaac55a0>]
Run the interpolation defining an isok:
isok = entry.isok(kx_pts, ky_pts, klabels)
Show the final results with the executed path on the isoenergy: sphinx_gallery_thumbnail_number = 6
fig, axs = plt.subplots(1, 2)
entry.isoenergy(0, 0.02).show(ax=axs[0])
isok.path_show(axs[0], 'k', 'k', xytext=(8, 8))
qmesh = isok.show(ax=axs[1])
fig.tight_layout()
fig.colorbar(qmesh)
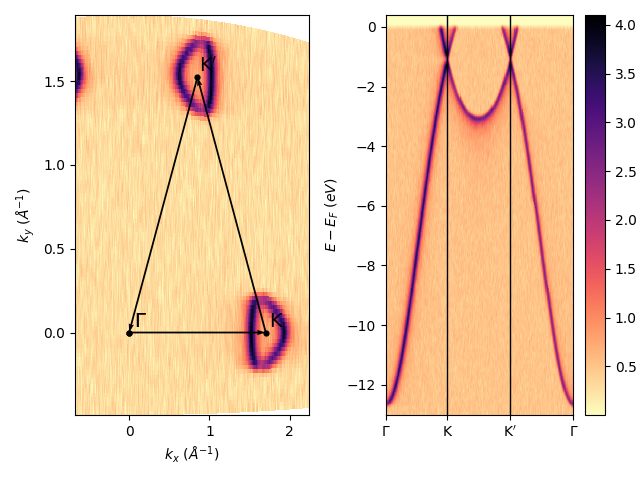
<matplotlib.colorbar.Colorbar object at 0x78b8ea7105b0>
Total running time of the script: (0 minutes 7.983 seconds)