Note
Go to the end to download the full example code.
The old way analysis¶
Old workflow for analyzing a deflector scan data. This workflow use the all the function in the most explicit way without using any entry method. This is not a recommended workflow but it can help on understanding what it is behind the entry methods.
Import the “fundamental” python libraries for a generic data analysis:
import numpy as np
import matplotlib.pyplot as plt
Import the navarp libraries:
from navarp.utils import navfile, fermilevel, navplt, ktransf, isocut
Load the data from a file:
file_name = r"nxarpes_simulated_cone.nxs"
entry = navfile.load(file_name)
instrument_name = simulated
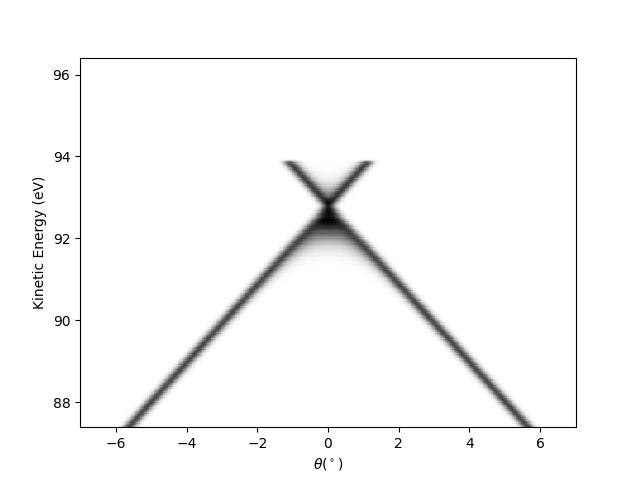
Plot a single slice Ex: scan = 0.5
scan = 0.5
scan_ind = np.argmin(abs(entry.scans-scan))
isoscan = isocut.maps_sum(scan, 0, entry.scans, entry.data)
qmisoscan = navplt.pimage(
entry.angles, entry.energies, isoscan, cmap='binary', style='tht_ekin')
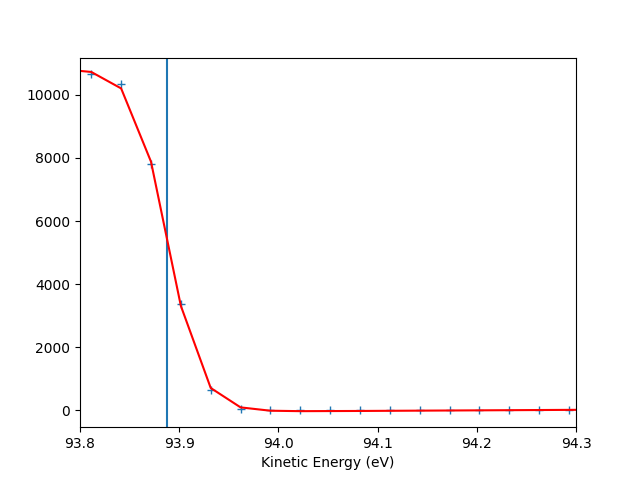
Fermi level determination
energy_range = [93.8, 94.3]
data_sum = np.sum(entry.data, axis=tuple([0, 1]))
popt = fermilevel.fit_efermi(entry.energies, data_sum, energy_range)
efermi, res = popt[0], popt[1]*4
fig, axfit = plt.subplots(1)
axfit.axvline(popt[0])
axfit.plot(entry.energies, data_sum, '+')
axfit.plot(entry.energies, fermilevel.fermi_fun(entry.energies, *popt), 'r-')
axfit.set_xlabel(r'Kinetic Energy (eV)')
axfit.set_xlim(energy_range)
dvis = data_sum[
(entry.energies >= energy_range[0]) &
(entry.energies <= energy_range[1])
]
dvis_min = dvis.min()
dvis_max = dvis.max()
dvis_delta = dvis_max - dvis_min
axfit.set_ylim(
dvis_min - dvis_delta*0.05,
dvis_max + dvis_delta*0.05
)
# Overwrite hv from the derived fermi level and the known work function
hv_from_file = np.copy(entry.hv)
entry.hv = np.array([efermi + entry.analyzer.work_fun])
print(
"hv = {:g} eV (from the file was {:g})".format(
np.squeeze(entry.hv), np.squeeze(hv_from_file))
)
print("Energy resolution = {:.0f} meV".format(res*1000))
/usr/local/lib/python3.10/site-packages/navarp/utils/fermilevel.py:67: RuntimeWarning: divide by zero encountered in divide
ddata_s_denergies = ddata_s_denergies/np.abs(data_sum)
/usr/local/lib/python3.10/site-packages/navarp/utils/fermilevel.py:67: RuntimeWarning: invalid value encountered in divide
ddata_s_denergies = ddata_s_denergies/np.abs(data_sum)
hv = 98.4881 eV (from the file was 100)
Energy resolution = 67 meV
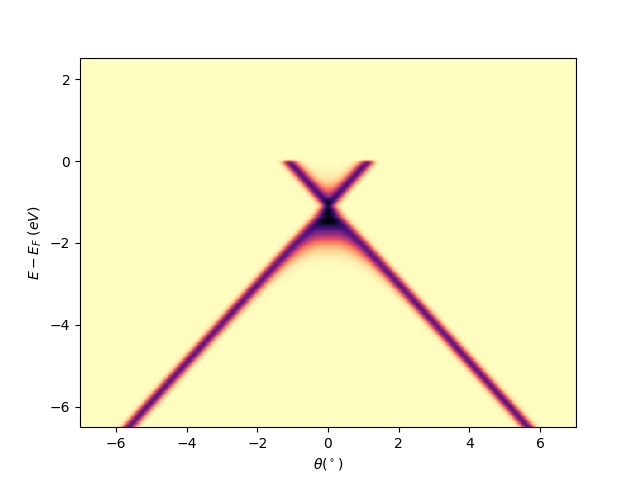
Plot a single slice with fermi level alignment Ex: scan = 0.5
scan = 0.5
scan_ind = np.argmin(abs(entry.scans-scan))
isoscan = isocut.maps_sum(scan, 0, entry.scans, entry.data)
qmisoscan = navplt.pimage(
entry.angles, entry.energies-efermi, isoscan,
cmap='magma_r', style='tht_eef')
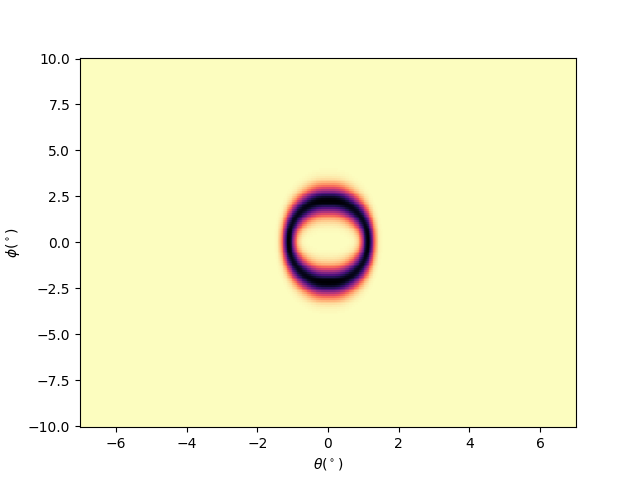
Plotting iso-energetic cut Ex: isoenergy cut at ekin = efermi
ekin = efermi
dekin = 0.005
isoev = isocut.maps_sum(ekin, dekin, entry.energies, entry.data)
qmisoev = navplt.pimage(
entry.angles, entry.scans, isoev,
cmap='magma_r', style='tht_phi', cmapscale='linear')
Total running time of the script: (0 minutes 1.264 seconds)